Sequence database setup: UniRef
|
Overview
UniRef, (UniProt Reference Clusters) provide clustered sets of sequences from the UniProt Knowledgebase (including isoforms). The seed sequences are the longest members of the cluster. There are three versions of UniRef: UniRef100, UniRef90, and UniRef50. UniRef100 is non-identical, while UniRef90 and UniRef50 are non-redundant at a sequence similarity level of 90% and 50% respectively. Searching with mass spectrometry data requires the exact sequence to be present in the database, so UniRef100 is the version to choose.
Using the database in Mascot 2.5 and later
Enable the predefined definition in Database Manager, which will download the required files automatically.
Using the database in Mascot 2.4
Enable the predefined definition in Database Manager to get the latest configuration. Downloading the files will fail, because Mascot 2.4 does not support HTTPS file downloads. Please download the FASTA file and required taxonomy files manually as described below.
Download (Mascot 2.3)
PIR:
https://ftp.uniprot.org/pub/databases/uniprot/uniref/uniref100/
EBI:
https://ftp.ebi.ac.uk/pub/databases/uniprot/uniref/uniref100/
Expasy:
https://ftp.uniprot.org/pub/databases/uniprot/current_release/uniref/uniref100/
The files are:
- Version info: uniref100.release_note
- Fasta file: uniref100.fasta.gz
Note that the XML file, uniref100.xml.gz, contains essentially the same information as the Fasta file. It is not a full text reference file.
To download SwissProt updates automatically in Mascot 2.3 and earlier, the relevant definition block in db_update.pl is UniRef100_Fasta_from_EBI.
Taxonomy (Mascot 2.3)
If you have Mascot 2.0 or earlier, add the following taxonomy definition to mascot.dat, changing the taxonomy block number so as to be consecutive with the existing blocks. If you have Mascot 2.1 or 2.2, you will need to update the existing taxonomy definition, because the database curators recently made changes to the fasta title syntax. Make a backup copy of mascot.dat, then use a text editor to make these changes. Note that the file must be saved as plain text, so be careful if using a word processor, and ensure the filename is not changed to mascot.dat.txt or something.
# TAXONOMY FOR UniRef
Taxonomy_12
Identifier UniRef Fasta
Enabled 1 # 0 to disable it
FromRefFile 0
ErrorLevel 0
SpeciesFiles NCBI:names.dmp
NodesFiles NCBI:nodes.dmp
DefaultRule NCBI, CHOP:W "Tax=\(.*\) RepID=" #…
end
The following taxonomy file is required:
https://ftp.ncbi.nlm.nih.gov/pub/taxonomy/taxdump.tar.gz
Remember that the taxonomy files go into the taxonomy directory, not into the sequence database directory. Also, these files need to be unpacked (using tar) as well as uncompressed.
Parse Rules (Mascot 2.3)
A typical UniRef Fasta title line is:
>UniRef100_Q4U9M9 104 kDa microneme/rhoptry antigen n=1 Tax=Theileria annulata RepID=104K_THEAN
The literal text, UniRef100_, should be dropped from the accession string, to make linking easier.
Accession from Fasta title: ">UniRef100_\([^ ]*\)"
Description from Fasta title: ">[^ ]* \(.*\)"
Configuration (Mascot 2.3 and earlier)
For this example, the fasta file was downloaded to C:\Inetpub\MASCOT\sequence\uniref100\current, decompressed using Gzip, and renamed to uniref100_9.6.fasta. Note that the rule numbers in your copy of mascot.dat may differ from those in the screen shot
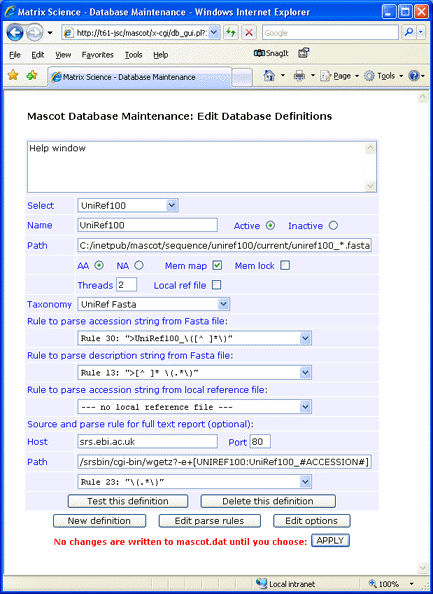
Update: It has become difficult to find an operating SRS server. Except for entries with UniParc identifiers, e.g. UPI00051B6503, annotation text for entries can be retrieved from UniProt using these settings:
Host: www.uniprot.org
Path: /uniprot/#ACCESSION#.txt
If you don’t require full text in a Mascot Protein View report, simply leave the Host, Port,
and Path fields blank and choose
— no full text report —
in the drop down list.
Always test a new definition before applying the changes to mascot.dat