Enzymes
Supported Reagents
| Name | Cleave | Don’t cleave | N or C term |
|---|---|---|---|
| Trypsin | KR | P | C |
| Trypsin/P | KR | C | |
| Arg-C | R | P | C |
| Asp-N | BD | N | |
| Asp-N_ambic | DE | N | |
| Chymotrypsin | FYWL | P | C |
| CNBr | M | C | |
| CNBr+Trypsin | M | C | |
| KR | P | C | |
| Formic_acid | D | C | |
| D | N | ||
| Lys-C | K | P | C |
| Lys-C/P | K | C | |
| LysC+AspN | K | P | C |
| DB | N | ||
| Lys-N | K | N | |
| NoCleave | see notes | ||
| PepsinA | FL | C | |
| semiTrypsin | see notes | ||
| TrypChymo | FYWLKR | P | C |
| TrypsinMSIPI | KR | P | C |
| J | C | ||
| J | N | ||
| TrypsinMSIPI/P | KR | C | |
| J | C | ||
| J | N | ||
| V8-DE | BDEZ | P | C |
| V8-E | EZ | P | C |
| None | see notes | ||
Up to 9 missed cleavage sites can be allowed, specified by the Missed Cleavages parameter.
"None" means that Mascot will search each protein sequence for every sub-sequence which meets the other search criteria. This usually means testing orders of magnitude more peptides than if trypsin cleavage had been specified. If the experimental data are from peptides that do not originate from an enzyme digest, such as MHC peptides, then "None" is the correct choice. Otherwise, ignoring the enzyme specificity greatly increases both the search time and, more importantly, the identity threshold, which will usually result in fewer significant matches in total. An error tolerant search is a much better way to identify the occasional non-specific cleavage product. "None" is never allowed for a Peptide Mass Fingerprint, where the specificity of an enzyme is essential.
"NoCleave" is used for top-down. It represents the absence of any enzyme and should not be confused with "None".
"semiTrypsin" means that Mascot will search for peptides that show tryptic specificity (KR not P) at one terminus, but where the other terminus may be a non-tryptic cleavage. This is a half-way house between choosing "Trypsin" and "None". It will only fail to find peptides that are non-specific at both ends.
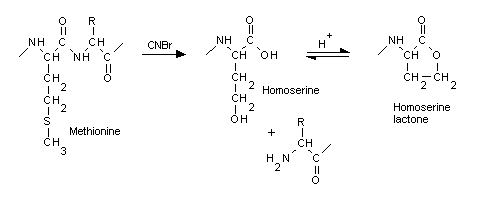
When a protein is cleaved by the commonly used enzymes, the new C terminus gains a hydroxyl group, while the new N terminus gains a hydrogen. The activity of cyanogen bromide (CNBr) is unusual in that cleaves on the C-terminal side of methionine, converting it to a homoserine. Under acidic conditions, homoserine generally cyclises to the lactone form, which does not require any terminating group. In Mascot, when CNBr is chosen as the cleavage agent, you also need to specify homoserine (Met->Hse) or homoserine lactone (Met->Hsl) as variable modifications.
Peptide Size Distribution
If we make the (fairly accurate) assumption that the proteins in a database can be treated as composed of truly random sequences of the 20 standard amino acids, then it is straightforward to calculate the peptide size distribution to be expected from any given enzyme.
In the following graph, peptide size distributions are plotted for limit digests of chymotrypsin (assumed to cleave at FWYLMH), trypsin, and CNBr:
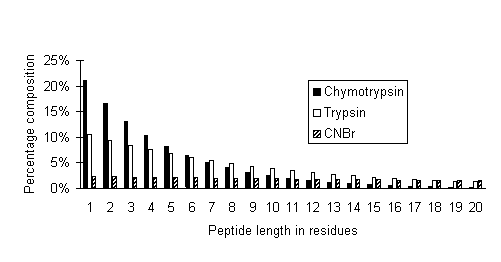
It can be seen that the high specificity cleavage reagent, CNBr, produces a relatively flat distribution of peptide sizes, ideal for fingerprinting by MALDI. In the case of the low specificity enzyme, chymotrypsin, less than 1% of all peptides are longer than 20 residues and 70% are between 1 and 5 residues. In a mass spectrum, peaks for these peptides would be crowded together into the low mass region, resulting in extensive overlapping.
Trypsin is somewhere in between, with 10% of peptides longer than 20 residues. Because of the relative scarcity of these larger peptides, it is the higher experimental mass values which provide greatest discrimination in a peptide mass fingerprint.